8. Examples of tool capabilities and applications¶
Additional examples of typical pipelines can be found in the Sentieon GitHub page in https://github.com/Sentieon/sentieon-scripts/tree/master/example_pipelines.
8.1. DNA pipeline example script¶
Below is an example of how to process a set of 2 fastq files for a sample and perform variant calling according to the Broad institute best practices described in https://www.broadinstitute.org/gatk/guide/best-practices.
#!/bin/sh
# *******************************************
# Script to perform DNA seq variant calling
# using a single sample with fastq files
# named 1.fastq.gz and 2.fastq.gz
# *******************************************
# Update with the fullpath location of your sample fastq
fastq_folder="/home/pipeline/samples"
fastq_1="1.fastq.gz"
fastq_2="2.fastq.gz" #If using Illumina paired data
sample="sample_name"
group="read_group_name"
platform="ILLUMINA"
# Update with the location of the reference data files
fasta="/home/regression/references/b37/human_g1k_v37_decoy.fasta"
dbsnp="/home/regression/references/b37/dbsnp_138.b37.vcf.gz"
known_Mills_indels="/home/regression/references/b37/Mills_and_1000G_gold_standard.indels.b37.vcf.gz"
known_1000G_indels="/home/regression/references/b37/1000G_phase1.indels.b37.vcf.gz"
# Update with the location of the Sentieon software package and license file
export SENTIEON_INSTALL_DIR=/home/release/sentieon-genomics-202010
export SENTIEON_LICENSE=/home/Licenses/Sentieon.lic
# Other settings
nt=$(nproc) #number of threads to use in computation, set to number of cores in the server
workdir="$PWD/test/DNAseq" #Determine where the output files will be stored
# ******************************************
# 0. Setup
# ******************************************
mkdir -p $workdir
logfile=$workdir/run.log
exec >$logfile 2>&1
cd $workdir
# ******************************************
# 1. Mapping reads with BWA-MEM, sorting
# ******************************************
#The results of this call are dependent on the number of threads used. To have number of threads independent results, add chunk size option -K 10000000
( $SENTIEON_INSTALL_DIR/bin/sentieon bwa mem -R "@RG\tID:$group\tSM:$sample\tPL:$platform" -t $nt -K 10000000 $fasta $fastq_folder/$fastq_1 $fastq_folder/$fastq_2 || echo -n 'error' ) | $SENTIEON_INSTALL_DIR/bin/sentieon util sort -r $fasta -o sorted.bam -t $nt --sam2bam -i -
# ******************************************
# 2. Metrics
# ******************************************
$SENTIEON_INSTALL_DIR/bin/sentieon driver -r $fasta -t $nt -i sorted.bam --algo MeanQualityByCycle mq_metrics.txt --algo QualDistribution qd_metrics.txt --algo GCBias --summary gc_summary.txt gc_metrics.txt --algo AlignmentStat --adapter_seq '' aln_metrics.txt --algo InsertSizeMetricAlgo is_metrics.txt
$SENTIEON_INSTALL_DIR/bin/sentieon plot GCBias -o gc-report.pdf gc_metrics.txt
$SENTIEON_INSTALL_DIR/bin/sentieon plot QualDistribution -o qd-report.pdf qd_metrics.txt
$SENTIEON_INSTALL_DIR/bin/sentieon plot MeanQualityByCycle -o mq-report.pdf mq_metrics.txt
$SENTIEON_INSTALL_DIR/bin/sentieon plot InsertSizeMetricAlgo -o is-report.pdf is_metrics.txt
# ******************************************
# 3. Remove Duplicate Reads. It is possible
# to mark instead of remove duplicates
# by ommiting the --rmdup option in Dedup
# ******************************************
$SENTIEON_INSTALL_DIR/bin/sentieon driver -t $nt -i sorted.bam --algo LocusCollector --fun score_info score.txt
$SENTIEON_INSTALL_DIR/bin/sentieon driver -t $nt -i sorted.bam --algo Dedup --rmdup --score_info score.txt --metrics dedup_metrics.txt deduped.bam
# ******************************************
# 5. Base recalibration
# ******************************************
$SENTIEON_INSTALL_DIR/bin/sentieon driver -r $fasta -t $nt -i deduped.bam --algo QualCal -k $dbsnp -k $known_Mills_indels -k $known_1000G_indels recal_data.table
$SENTIEON_INSTALL_DIR/bin/sentieon driver -r $fasta -t $nt -i deduped.bam -q recal_data.table --algo QualCal -k $dbsnp -k $known_Mills_indels -k $known_1000G_indels recal_data.table.post
$SENTIEON_INSTALL_DIR/bin/sentieon driver -t $nt --algo QualCal --plot --before recal_data.table --after recal_data.table.post recal.csv
$SENTIEON_INSTALL_DIR/bin/sentieon plot QualCal -o recal_plots.pdf recal.csv
# ******************************************
# 6b. HC Variant caller
# ******************************************
$SENTIEON_INSTALL_DIR/bin/sentieon driver -r $fasta -t $nt -i deduped.bam -q recal_data.table --algo Haplotyper -d $dbsnp --emit_conf=30 --call_conf=30 output-hc.vcf.gz
# ******************************************
# 5b. ReadWriter to output recalibrated bam
# This stage is optional as variant callers
# can perform the recalibration on the fly
# using the before recalibration bam plus
# the recalibration table
# ******************************************
$SENTIEON_INSTALL_DIR/bin/sentieon driver -r $fasta -t $nt -i deduped.bam -q recal_data.table --algo ReadWriter recaled.bam
8.2. Working with multiple input files¶
Typically, there will be two occasions when you will be having multiple input files that need to be processed together: a single sample has been sequenced in multiple lanes, or you are processing multiple samples from a cohort.
8.2.1. Working with multiple input files from the same sample¶
When you have more than one set of input fastq files for the sample, you should perform an individual alignment stage for each set of input fastq files, and then use the multiple sorted BAM files as input of the next stage. The next stage will take care of merging all the BAM files into one. You need to make sure that each set of input fastq files are aligned using the same SM sample name, but a different RG readgroup name, so that the rest of the stages properly deal with the different data.
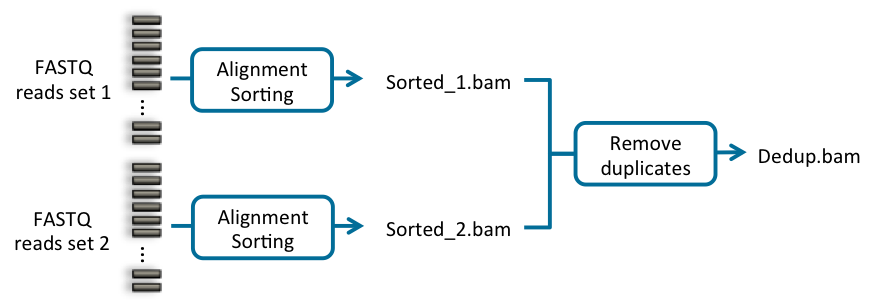
Fig. 8.1 Bioinformatics pipeline using multiple input files from the same sample
#Run alignment for 1st input file set
(sentieon bwa mem -R '@RG\tID:GROUP_NAME_1 \tSM:SAMPLE_NAME \tPL:PLATFORM' \
-t NUMBER_THREADS REFERENCE SAMPLE_1 [SAMPLE_1_2] || echo -n 'error' ) \
| sentieon util sort -o SORTED_BAM_1 -t NUMBER_THREADS --sam2bam -i -
#Run alignment for 2nd input file set
(sentieon bwa mem -R '@RG\tID:GROUP_NAME_2 \tSM:SAMPLE_NAME \tPL:PLATFORM' \
-t NUMBER_THREADS REFERENCE SAMPLE_2 [SAMPLE_2_2] || echo -n 'error' \
sentieon util sort -o SORTED_BAM_2 -t NUMBER_THREADS --sam2bam -i -
#Run dedup on both BAM files
sentieon driver -t NUMBER_THREADS -i SORTED_BAM_1 -i SORTED_BAM_2 \
--algo LocusCollector --fun score_info SCORE_TXT
sentieon driver -t NUMBER_THREADS -i SORTED_BAM_1 -i SORTED_BAM_2 \
--algo Dedup --rmdup --score_info SCORE_TXT --metrics DEDUP_METRIC_TXT DEDUPED_BAM
Alternatively you can input the results of multiple BWA alignments onto a single util sort command to merge the results onto a single sorted bam files as soon as possible. You still need to make sure that each set of input fastq files are aligned using the same SM sample name, but a different RG readgroup name, so that the rest of the stages properly deal with the different data.
#Run alignment for both input file sets sequentially
(sentieon bwa mem -R '@RG\tID:GROUP_NAME_1\tSM:SAMPLE_NAME\tPL:PLATFORM' \
-t NUMBER_THREADS REFERENCE SAMPLE_1 [SAMPLE_1_2] && sentieon bwa mem -R \
'@RG\tID:GROUP_NAME_2 \tSM:SAMPLE_NAME \tPL:PLATFORM' \
-t NUMBER_THREADS REFERENCE SAMPLE_2 [SAMPLE_2_2] || echo -n 'error' ) \
| sentieon util sort -o SORTED_COMBINED_BAM -t NUMBER_THREADS --sam2bam -i -
#Run dedup on combined BAM file
sentieon driver -t NUMBER_THREADS -i SORTED_COMBINED_BAM_1 \
--algo LocusCollector --fun score_info SCORE_TXT
sentieon driver -t NUMBER_THREADS -i SORTED_COMBINED_BAM_1 \
--algo Dedup --rmdup --score_info SCORE_TXT --metrics DEDUP_METRIC_TXT DEDUPED_BAM
8.2.2. Working with multiple input files from different samples¶
Processing multiple samples from a cohort together, also known as joint variant calling, can be done in two ways:
- Process each sample individually and use the Haplotyper algorithm
with option
--emit_mode gvcfto create a GVCF file containing additional information, then process all GVCF using the GVCFtyper algorithm. This method allows for easy and fast reprocessing after additional samples have been processed.
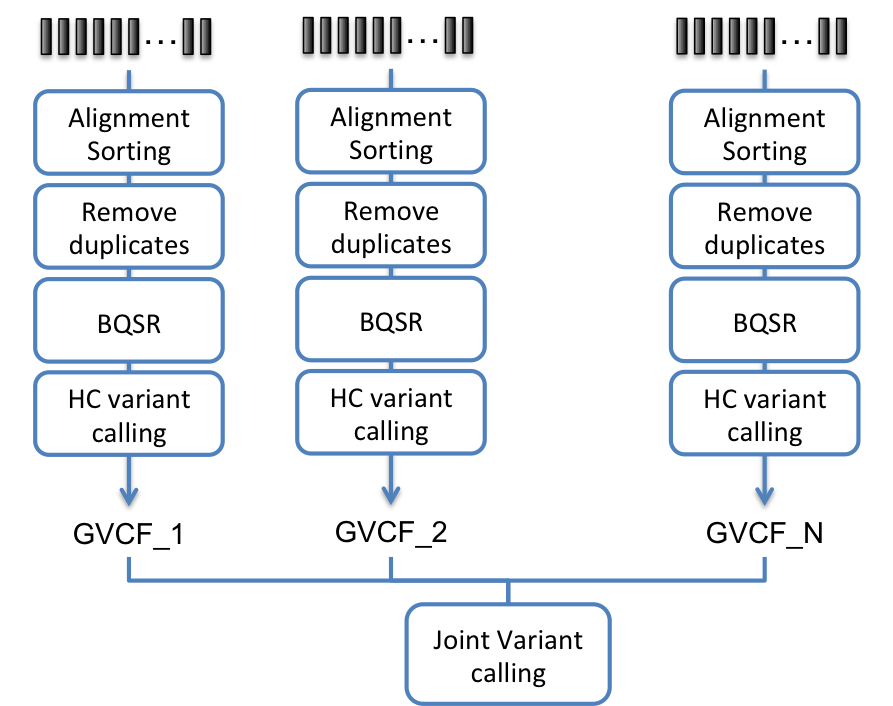
Fig. 8.2 Bioinformatics pipeline for joint calling using Haplotyper in GVCF emit mode and GVCFtyper
#Process all samples independently until GVCF
for sample in s1 s2 s3; do
(sentieon bwa mem -R '@RG\tID:GROUP_${sample}\tSM:${sample}\tPL:PLATFORM' \
-t NUMBER_THREADS REFERENCE ${sample}_FASTQ_1 ${sample}_FASTQ_2 || echo -n \
'error') | sentieon util sort -o ${sample}_SORTED_BAM -t NUMBER_THREADS \
--sam2bam -i -
sentieon driver -t NUMBER_THREADS -i ${sample}_SORTED_BAM \
--algo LocusCollector --fun score_info ${sample}_SCORE_TXT
sentieon driver -t NUMBER_THREADS -i SORTED_BAM_1 -i SORTED_BAM_2 \
--algo Dedup --rmdup --score_info ${sample}_SCORE_TXT ${sample}_DEDUPED_BAM
sentieon driver -r REFERENCE -t NUMBER_THREADS -i ${sample}_DEDUPED_BAM \
--algo QualCal -k $known_sites ${sample}_RECAL_DATA_TABLE
sentieon driver -r REFERENCE -t NUMBER_THREADS -i ${sample}_DEDUPED_BAM \
-q ${sample}_RECAL_DATA_TABLE --algo Haplotyper --emit_mode gvcf \
${sample}_VARIANT_GVCF
done
#Run joint variant calling
sentieon driver -r REFERENCE --algo GVCFtyper -v s1_VARIANT_GVCF \
-v s2_VARIANT_GVCF -v s3_VARIANT_GVCF VARIANT_VCF
- Process each sample individually and create either a recalibrated BAM file or a deduped BAM and a recalibration table for each sample, then process all files using the Haplotyper algorithm.
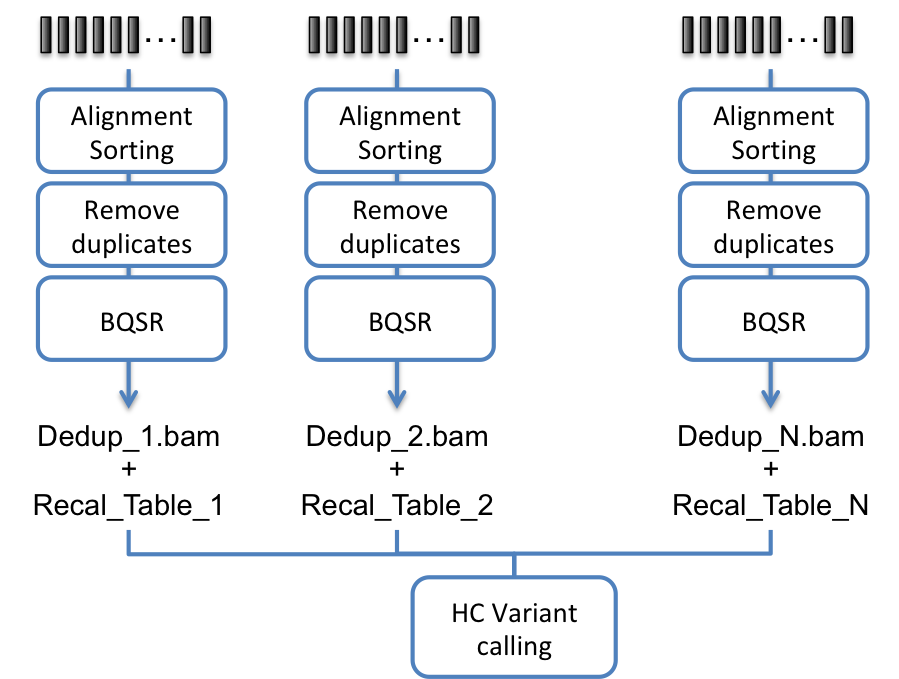
Fig. 8.3 Bioinformatics pipeline for joint calling using Haplotyper and multiple BAM files
#Process all samples independently until BQSR
for sample in s1 s2 s3; do
(sentieon bwa mem -R '@RG\tID:GROUP_${sample}\tSM:${sample}\tPL:PLATFORM' \
-t NUMBER_THREADS REFERENCE ${sample}_FASTQ_1 ${sample}_FASTQ_2 || echo -n \
'error' )| sentieon util sort -o ${sample}_SORTED_BAM -t NUMBER_THREADS \
--sam2bam -i -
sentieon driver -t NUMBER_THREADS -i ${sample}_SORTED_BAM \
--algo LocusCollector --fun score_info ${sample}_SCORE_TXT
sentieon driver -t NUMBER_THREADS -i SORTED_BAM_1 -i SORTED_BAM_2 \
--algo Dedup --rmdup --score_info ${sample}_SCORE_TXT ${sample}_DEDUPED_BAM
sentieon driver -r REFERENCE -t NUMBER_THREADS -i ${sample}_DEDUPED_BAM \
--algo QualCal -k $known_sites ${sample}_RECAL_DATA_TABLE
done
#Run joint variant calling
sentieon driver -t NUMBER_THREADS -i s1_DEDUPED_BAM -q s1_RECAL_DATA_TABLE \
-i s2_DEDUPED_BAM -q s2_RECAL_DATA_TABLE -i s3_DEDUPED_BAM \
-q s3_RECAL_DATA_TABLE --algo Haplotyper VARIANT_VCF
8.3. Supported annotations in Haplotyper and Genotyper¶
Both Haplotyper and Genotyper germline variant calling methods accept
the option --annotation to output additional annotations to the result
VCF. The following are the possible annotations supported:
- AC: Allele count in genotypes, for each ALT allele, in the same order as listed
- AF: Allele frequency, for each ALT allele, in the same order as listed
- AN: Total number of alleles in called genotypes
- BaseQRankSum: Z-score from Wilcoxon rank sum test of Alt Vs. Ref base qualities
- ClippingRankSum: Z-score From Wilcoxon rank sum test of Alt vs. Ref number of hard clipped bases
- DP: Combined depth across samples
- ExcessHet: Phred-scaled p-value for exact test of excess heterozygosity
- FS: Phred-scaled p-value using Fisher's exact test to detect strand bias
- InbreedingCoeff: Inbreeding coefficient as estimated from the genotype likelihoods per-sample when compared against the Hardy-Weinberg expectation
- MLEAC: Maximum likelihood expectation (MLE) for the allele counts, for each ALT allele, in the same order as listed
- MLEAF: Maximum likelihood expectation (MLE) for the allele frequency, for each ALT allele, in the same order as listed
- MQ: RMS mapping quality
- MQ0: Number of MAPQ == 0 reads
- MQRankSum: Z-score From Wilcoxon rank sum test of Alt vs. Ref read mapping qualities
- QD: Variant Confidence/Quality by Depth
- RAW_MQ: Raw data for RMS mapping quality
- ReadPosRankSum: Z-score from Wilcoxon rank sum test of Alt vs. Ref read position bias
- SOR: Symmetric Odds Ratio of 2x2 contingency table to detect strand bias
- SAC: Number of reads on the forward and reverse strand supporting each allele (including reference)
- AS_BaseQRankSum: Allele specific
- AS_FS: Allele specific Phred-scaled p-value using Fisher's exact test to detect strand bias
- AS_InbreedingCoeff: Allele specific Inbreeding coefficient as estimated from the genotype likelihoods per-sample when compared against the Hardy-Weinberg expectation
- AS_MQRankSum: Allele specific Z-score From Wilcoxon rank sum test of Alt vs. Ref read mapping qualities
- AS_QD: Allele specific Variant Confidence/Quality by Depth
- AS_MQ: Allele specific RMS mapping quality
- AS_ReadPosRankSum: Allele specific Z-score from Wilcoxon rank sum test of Alt vs. Ref read position bias
- AS_SOR: Allele specific Symmetric Odds Ratio of 2x2 contingency table to detect strand bias
8.4. Removing reads after alignment with low mapping quality¶
Below is an example of how to remove reads with low mapping quality after alignment, so that they do not take space in the output BAM file.
sentieon bwa mem -R '@RG\tID:GROUP_NAME_1 \tSM:SAMPLE_NAME \tPL:PLATFORM' \
-t NUMBER_THREADS REFERENCE SAMPLE_1 [SAMPLE_2] \
|samtools view -h -q MAP_QUALITY_THRESHOLD - \
| sentieon util sort -o SORTED_BAM_1 -t NUMBER_THREADS --sam2bam -i -
8.5. Performing Dedup to mark primary and non-primary reads¶
The standard 2 pass dedup will ignore non-primary reads and never change their duplicate flag.
Below is an example of how to perform dedup to mark both primary and non-primary reads:
sentieon driver -t NUMBER_THREADS -i SORTED_BAM \
--algo LocusCollector --fun score_info SCORE_TXT
sentieon driver -t NUMBER_THREADS -i SORTED_BAM \
--algo Dedup --score_info SCORE_TXT --metrics DEDUP_METRIC_TXT \
-- output_dup_read_name DUPLICATED_READNAMES_TXT
sentieon driver -t NUMBER_THREADS -i SORTED_BAM \
--algo Dedup --rmdup --dup_read_name DUPLICATED_READNAMES_TXT DEDUPED_BAM
8.6. Pipeline modifications when using data with quantized quality scores¶
When your input data has quantized base quality scores, there is a high
chance that the base quality reported in the input FASTQ is much higher
than the empirical base quality; in this case, it is possible that the
variant calling step may take longer to process the data, as low quality
bases that would not have been taken into account in the local assembly
will cause the generation of many irrelevant low-quality haplotypes.
This issue should be addressed by running the Base Quality Score
Recalibration step of the pipeline; alternatively, you can modify
the default value of the min_base_qual argument of Haplotyper, or
TNhaplotyper to reduce the impact: add the option --min_base_qual 15
to your command line to prevent this issue.
8.7. Modify RG information on BAM files when both Tumor and Normal inputs have the same RGID¶
If you input multiple different BAM files containing readgroups with the same ID but
different attributes, the Sentieon® tools will error out. This can happen when in
TNseq® and TNscope® the tumor and normal sample BAM files have an RG ID of “1”.
In order to work around this, you can use the --replace_rg functionality to modify
the RG information.
#first lane tumor, RGID incorrectly set to uninformative 1
sentieon bwa mem -R '@RG\tID:1\tSM:TUMOR_SM\tPL:PLATFORM' -t NT \
REFERENCE TUMOR_1_1.fq.gz TUMOR_1_2.fq.gz | sentieon util sort \
-o TUMOR_1.bam -t NT --sam2bam -i -
#second lane tumor, RGID incorrectly set to uninformative 1
sentieon bwa mem -R '@RG\tID:1\tSM:TUMOR_SM\tPL:PLATFORM' -t NT \
REFERENCE TUMOR_2_1.fq.gz TUMOR_2_2.fq.gz | sentieon util sort \
-o TUMOR_2.bam -t NT --sam2bam -i -
#normal, RGID incorrectly set to uninformative 1
sentieon bwa mem -R '@RG\tID:1\tSM:NORMAL_SM\tPL:PLATFORM' -t NT \
REFERENCE TUMOR_1.fq.gz TUMOR_2.fq.gz | sentieon util sort \
-o NORMAL.bam -t NT --sam2bam -i -
#using replace_rg argument to modify the RGID and make them unique
#each replace_RG argument will only affect the following input BAM file
sentieon driver -r REFERENCE \
--replace_rg 1='ID:T_1\tSM:TUMOR\tPL:PLATFORM' -i TUMOR_1.bam \
--replace_rg 1='ID:T_2\tSM:TUMOR\tPL:PLATFORM' -i TUMOR_2.bam \
--replace_rg 1='ID:N\tSM:NORMAL\tPL:PLATFORM'-i NORMAL.bam \
--algo TNscope --tumor_sample TUMOR --normal_sample NORMAL OUT_TN_VCF
8.8. Running the license server (LICSRVR) as a system service¶
8.8.1. Running the license server as a system service using sysvinit¶
If your system follows the traditional System V init startup scripts you can setup the license server to be automatically started in your system by running the following commands as root:
Create and customize the configuration file; the configuration file is typically /etc/sysconfig/licsrvr; however, in Ubuntu the configuration file will be /etc/default/licsrvr. Below is an example of the configuration file, for the recommended setup using the
sentieonusername:/home/sentieon/release/latestis a symlink to the installation directory of the latest Sentieon® software package/home/sentieon/licsrvris a folder to run the licsrvr service/home/sentieon/licsrvr/licsrvr.licis the Sentieon® license file
licsrvr="/home/sentieon/release/latest/bin/sentieon licsrvr" licfile=/home/sentieon/licsrvr/licsrvr.lic logfile=/home/sentieon/licsrvr/licsrvr.log
Install the license server startup script into the /etc/init.d directory. The startup script is included with the release package.
install -m 0755 $SENTIEON_INSTALL_DIR/doc/licsrvr.sh /etc/init.d/licsrvr
Install and enable the service. Depending on your system, you will run different commands:
- If your system has Linux Standard Base Core Specifications installed, execute the system init script installation script.
/usr/lib/lsb/install_initd /etc/init.d/licsrvr
If your system does not have the lsb conformance packages installed, use the
chkconfigcommand to enable the service.chkconfig --add licsrvr chkconfig licsrvr onFor Ubuntu and Debian systems, if you do not have the lsb/install_initd binary and choose not to install the lsb-core package, use the
update-rc.dcommand to install and enable the service.update-rc.d licsrvr defaults update-rc.d licsrvr enable
You can use the
servicecommand to start/stop/restart/check status of the service.service licsrvr [start|stop|restart|status]
8.8.2. Running the license server as a system service using systemd¶
You can use the systemd system and service capabilities of your OS to setup the license server to be automatically started in your system. To do so, run the following commands as root:
- If you are using the licsrvr.service license server startup script found in the
doc folder of the Sentieon® software release package, you will need to create
the necessary files that the script requires,
including using the
sentieonusername:/home/sentieon/release/latestis a symlink to the installation directory of the latest Sentieon® software package/home/sentieon/licsrvris a folder to run the licsrvr service/home/sentieon/licsrvr/licsrvr.licis the Sentieon® license file
Alternatively, you can edit the license server startup script to point to your specific username and/or file location information.
Install the license server startup script into the /etc/systemd/system directory.
install -m 0644 $SENTIEON_INSTALL_DIR/doc/licsrvr.service /etc/systemd/system
Run the following command to enable automatic start of the license server on the computer start:
systemctl enable licsrvr.serviceYou can use the
systemctlcommand to manually start and stop the service.systemctl start licsrvr.service systemctl stop licsrvr.service
