4. Typical usage for TNseq¶
Another of the typical uses of Sentieon Genomics software is to perform the bioinformatics pipeline for Tumor-Normal analysis recommended in the Broad institute Somatic short variant discovery (SNVs + Indels). Fig. 4.1 illustrates such a typical bioinformatics pipeline.
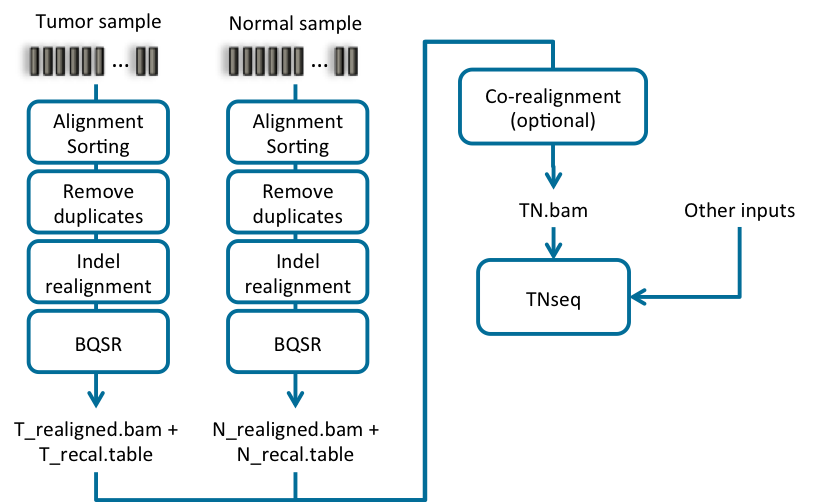
Fig. 4.1 Recommended bioinformatics pipeline for Tumor-Normal analysis
4.1. General¶
In this bioinformatics pipeline you will need the following inputs:
- The FASTA file containing the nucleotide sequence of the reference genome corresponding to the sample you will analyze.
- Two sets of FASTQ files containing the nucleotide sequence of the sample to be analyzed, one for the tumor sample and one for the matched normal sample. These files contain the raw reads from the DNA sequencing. The software supports inputting FASTQ files compressed using GZIP. The software only supports files containing quality scores in Sanger format (Phred+33).
- (Optional) The Single Nucleotide Polymorphism database (dbSNP) data that you want to include in the pipeline. The data is used in the form of a VCF file.
- (Optional) Multiple collections of known sites that you want to include in the pipeline. The data is used in the form of a VCF file.
You can also include in the pipeline the following optional inputs that will help the algorithms detect artifacts and remove false positives:
- Panel of normal VCF: list of common errors that appear as variants from multiple unrelated normal samples. The contents of this file will be used to identify variants that are more likely to be germline variants, and filter them as such.
- Cosmic VCF: data from the Catalogue of Somatic Mutations in Cancer (COSMIC) representing a list of known tumor related variants. The contents of this file will be used to reduce the germline risk factor of the variants. You need to use the same COSMIC file as the one used to generate the Panel of Normal VCF.
If you do not have accessed to a normal (non tumor) sample matched to the tumor sample, the Panel of Normal VCF and the Cosmic VCF inputs are highly recommended.
The following steps compose the typical bioinformatics pipeline for a tumor-normal matched pair:
- Independently pre-process the Tumor and Normal samples using a DNA
seq pipeline like the one introduced in Section 2, with the following stages:
- Map reads to reference; you need to make sure that the SM sample tag is different between the tumor and the normal samples, as you will need it as an argument in the somatic variant calling.
- Calculate data metrics.
- Remove duplicates.
- Indel realignment.
- Base quality score recalibration (BQSR).
- Indel co-realignment: This step performs a local realignment around indels for the combined data of both tumor and normal sample.
- Somatic variant calling on the co-realigned BAM or on the two individual bam: This step identifies the sites where the cancer genome data displays somatic variations relative to the normal genome, and calculates genotypes at that site.
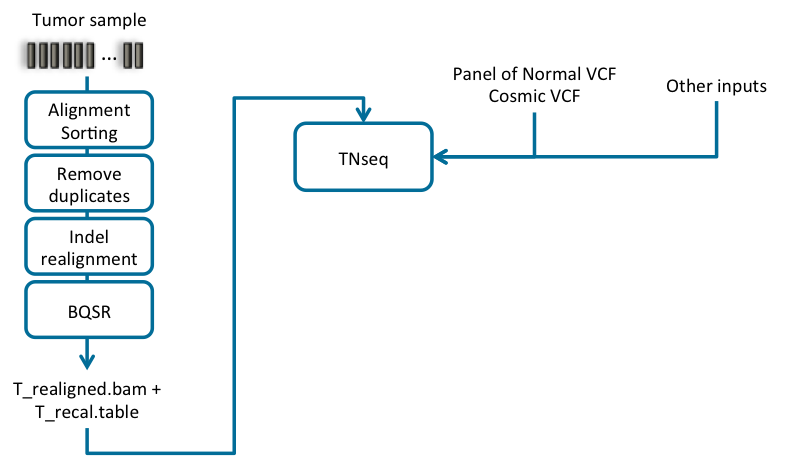
Fig. 4.2 Recommended bioinformatics pipeline for Tumor only analysis
The following steps compose the typical bioinformatics pipeline for a tumor sample without normal matched sample (Fig. 4.2):
- Pre-process the Tumor sample using a DNA seq pipeline like the one introduced in Section 2, with the
following stages:
- Map reads to reference.
- Calculate data metrics.
- Remove duplicates.
- Indel realignment.
- Base quality score recalibration (BQSR).
- (optional) Apply recalibration and create the recalibrated BAM file.
- Variant calling on the tumor sample using information from a generic panel of normal samples: This step identifies the sites where the cancer genome data displays somatic variations relative to the panel of normal genome, and calculates genotypes at that site.
4.2. Step by step usage¶
For the mapping, duplicate removal, indel realignment and base quality score recalibration stages, please refer to Section 2.2 for detailed usage instructions.
4.2.1. Indel co-realignment¶
A single command is run to perform local realignment around indels on the combination of both Tumor and Normal BAM files after pre-processing.
sentieon driver -t NUMBER_THREADS -r REFERENCE -i TUMOR_REALIGN_BAM \
-q TUMOR_RECAL_DATA.TABLE -i NORMAL_REALIGN_BAM -q NORMAL_RECAL_DATA.TABLE \
--algo Realigner [-k KNOWN_SITES] COREALIGNED_BAM
Alternatively, you can use already recalibrated BAM files to achieve the same results.
sentieon driver -t NUMBER_THREADS -r REFERENCE -i TUMOR_RECALIBRATED_BAM \
-i NORMAL_RECALIBRATED_BAM --algo Realigner [-k KNOWN_SITES] COREALIGNED_BAM
The following inputs are required for the command:
- NUMBER_THREADS: the number of computer threads that will be used in the calculation. We recommend that the number does not exceed the number of computing cores available in your system.
- REFERENCE: the location of the reference FASTA file. You should make sure that the reference is the same as the one used in the mapping stage.
- TUMOR_REALIGNED_BAM: the location of the pre-processed BAM file after individual realignment for the TUMOR sample.
- TUMOR_RECAL_DATA.TABLE: the location where the BQSR stage for the TUMOR sample stored the result.
- NORMAL_REALIGNED_BAM: the location of the pre-processed BAM file after individual realignment for the NORMAL sample.
- NORMAL_RECAL_DATA.TABLE: the location where the BQSR stage for the NORMAL sample stored the result.
- TUMOR_RECALIBRATED_BAM: the location of the pre-processed BAM file after recalibration for the TUMOR sample.
- NORMAL_RECALIBRATED_BAM: the location of the pre-processed BAM file after recalibration for the NORMAL sample.
- COREALIGNED_BAM: the location and filename of the realigned BAM output file. A corresponding index file (.bai) will be created.
The following inputs are optional for the command:
- KNOWN_SITES: the location of the VCF file used as a set of known
sites. You can include multiple collections of known sites by
repeating the
-k KNOWN_SITESoption.
4.2.2. Variant discovery with matched normal sample¶
A single command is run to call variants on the tumor-normal matched pair.
sentieon driver -t NUMBER_THREADS -r REFERENCE -i COREALIGNED_BAM \
--algo TNhaplotyper --tumor_sample TUMOR_SAMPLE_NAME \
--normal_sample NORMAL_SAMPLE_NAME [--dbsnp DBSNP] OUT_TN_VCF
You can also use the TNhaplotyper2 algorithm to call variants, in which case two commands are used:
sentieon driver -t NUMBER_THREADS -r REFERENCE -i COREALIGNED_BAM \
--algo TNhaplotyper2 --tumor_sample TUMOR_SAMPLE_NAME \
--normal_sample NORMAL_SAMPLE_NAME TMP_OUT_TN_VCF
sentieon tnhapfilter --tumor_sample TUMOR_SAMPLE_NAME \
--normal_sample NORMAL_SAMPLE_NAME -v TMP_OUT_TN_VCF OUT_TN_VCF
Alternatively, you can use TNsnv to call SNVs only:
sentieon driver -t NUMBER_THREADS -r REFERENCE -i COREALIGNED_BAM \
--algo TNsnv --tumor_sample TUMOR_SAMPLE_NAME \
--normal_sample NORMAL_SAMPLE_NAME [--dbsnp DBSNP] \
[--call_stats_out OUT_CALL_STATS] OUT_TN_VCF
Alternatively, if you did not run co-realignment, you can use the before co-realignment BAM files to perform the somatic calling:
sentieon driver -t NUMBER_THREADS -r REFERENCE -i TUMOR_REALIGN_BAM \
-q TUMOR_RECAL_DATA.TABLE -i NORMAL_REALIGN_BAM \
-q NORMAL_RECAL_DATA.TABLE --algo TNhaplotyper \
--tumor_sample TUMOR_SAMPLE_NAME --normal_sample NORMAL_SAMPLE_NAME \
[--dbsnp DBSNP] OUT_TN_VCF
The following inputs are required for the command:
- NUMBER_THREADS: the number of computer threads that will be used in the calculation. We recommend that the number does not exceed the number of computing cores available in your system.
- REFERENCE: the location of the reference FASTA file. You should make sure that the reference is the same as the one used in the mapping stage.
- COREALIGNED_BAM: the location and filename of the co-realigned BAM file from co-realignment stage.
- TUMOR_SAMPLE_NAME: sample name used for tumor sample in Map reads to reference stage.
- NORMAL_SAMPLE_NAME: sample name used for normal sample in Map reads to reference stage.
- TMP_OUT_TN_VCF: the location and file name of the output file from TNhaplotyper2; this is a temporary file.
- OUT_TN_VCF: the location and file name of the output file containing the variants.
The following inputs are optional for the command:
- DBSNP: the location of the Single Nucleotide Polymorphism database (dbSNP). The variants in the dbSNP will be more likely to be marked as germline as they require more evidence of absence in the normal. You can only use one dbSNP file.
- OUT_CALL_STATS: the location and file name of the output file containing the call stats of the discovered variants. The file is in text format. This argument is only needed when using TNsnv.
4.2.3. Step by step usage when missing a matched normal sample¶
When missing a matched normal sample, you will need to replace the matched normal sample with a generic Panel of Normal (PoN) file generated following the instructions in the next section.
A single command is run to call variants on the tumor only sample.
sentieon driver -t NUMBER_THREADS -r REFERENCE -i TUMOR_REALIGNED_BAM \
-q TUMOR_RECAL_DATA.TABLE --algo TNhaplotyper \
--tumor_sample TUMOR_SAMPLE_NAME --pon PANEL_OF_NORMAL_VCF \
--cosmic COSMIC_DB_VCF [--dbsnp DBSNP] OUT_TN_VCF
Alternatively, you can use already recalibrated BAM files to achieve the same results.
sentieon driver -t NUMBER_THREADS -r REFERENCE -i TUMOR_RECALIBRATED_BAM \
--algo TNhaplotyper --tumor_sample TUMOR_SAMPLE_NAME \
--pon PANEL_OF_NORMAL_VCF --cosmic COSMIC_DB_VCF \
[--dbsnp DBSNP] OUT_TN_VCF
You can also use the TNhaplotyper2 algorithm to call variants on the tumor sample using a PoN, in which case you use the following two commands:
sentieon driver -t NUMBER_THREADS -r REFERENCE -i TUMOR_REALIGNED_BAM \
-q TUMOR_RECAL_DATA.TABLE --algo TNhaplotyper2 \
--tumor_sample TUMOR_SAMPLE_NAME \
--pon PANEL_OF_NORMAL_VCF TMP_OUT_TN_VCF
sentieon tnhapfilter --tumor_sample TUMOR_SAMPLE_NAME \
-v TMP_OUT_TN_VCF OUT_TN_VCF
Bear in mind that when using TNhaplotyper2 without a normal sample, all variants in the output VCF file will be marked with the germline_risk FILTER by default, in addition to any other FILTER; these variants are considered to be potentially germline because of the lack of matched normal. As such, you may want to remove the germline_risk FILTER using the following BCFtools command:
BCF=/path_to_bcftools
$BCF/bcftools annotate -x FILTER/germline_risk OUT_TN_VCF OUT_TN_FILTER_VCF
The following inputs are required for the commands:
- NUMBER_THREADS: the number of computer threads that will be used in the calculation. We recommend that the number does not exceed the number of computing cores available in your system.
- REFERENCE: the location of the reference FASTA file. You should make sure that the reference is the same as the one used in the mapping stage.
- TUMOR_REALIGNED_BAM: the location of the pre-processed BAM file after realignment for the TUMOR sample.
- TUMOR_RECAL_DATA.TABLE: the location where the BQSR stage for the TUMOR sample stored the result.
- TUMOR_RECALIBRATED_BAM: the location of the pre-processed BAM file after recalibration for the TUMOR sample.
- TUMOR_SAMPLE_NAME: sample name used for tumor sample in Map reads to reference stage.
- PANEL_OF_NORMAL_VCF: the location and name of panel of normal VCF file.
- COSMIC_VCF: the location and name of the cosmic database VCF file. This file is only required for a PoN to be used with TNsnv or TNhaplotyper.
- OUT_TN_VCF: the location and file name of the output file containing the variants.
The following inputs are optional for the command:
- DBSNP: the location of the Single Nucleotide Polymorphism database (dbSNP). The variants in the dbSNP will be more likely to be marked as germline as they require more evidence of absence in the normal. You can only use one dbSNP file.
4.2.4. Generating a Panel of Normal VCF file¶
In order to generate your own Panel of Normal VCF file to use with TNhaplotyper, you will need to run the following command on every normal sample you want to use in the panel:
sentieon driver -t NUMBER_THREADS -r REFERENCE -i NORMAL_RECALIBRATED_BAM \
--algo TNhaplotyper --detect_pon \
--cosmic COSMIC_VCF [--dbsnp DBSNP] OUT_NORMAL_VCF
If you need to create a Panel of Normal VCF file to use with TNhaplotyper2, you will need to run the following command on every normal sample you want to use in the panel:
sentieon driver -t NUMBER_THREADS -r REFERENCE -i NORMAL_RECALIBRATED_BAM \
--algo TNhaplotyper2 --tumor_sample NORMAL_SAMPLE_NAME OUT_NORMAL_VCF
We recommend that you create the panel of normal file with the corresponding algorithm that you plan to use for the somatic mutation calling. If you plan to use TNsnv for the calling, you should use TNsnv for the generation of the panel of normal.
The following inputs are required for the commands:
- NUMBER_THREADS: the number of computer threads that will be used in the calculation. We recommend that the number does not exceed the number of computing cores available in your system.
- REFERENCE: the location of the reference FASTA file. You should make sure that the reference is the same as the one used in the mapping stage.
- NORMAL_RECALIBRATED_BAM: the location of the pre-processed BAM file for the normal sample after Sentieon Readwrite stage.
- COSMIC_VCF: the location and name of the cosmic database VCF file. This file is only required for a PoN to be used with TNsnv or TNhaplotyper.
- OUT_NORMAL_VCF: the location and name of the output VCF file containing the relevant variants for the input normal sample.
- NORMAL_SAMPLE_NAME: sample name used for normal sample in Map reads to reference stage.
The following inputs are optional for the command:
- DBSNP: the location of the Single Nucleotide Polymorphism database (dbSNP) that will be used to label known variants. You can only use one dbSNP file.
After you have generated all VCF files you want to include in the panel, you need to merge them into a single Panel of Normal VCF. You can use bcftools for that purpose:
BCF=/path_to_bcftools
export BCFTOOLS_PLUGINS=$BCF/plugins
DIR=/path_to_normal_vcf_file
$BCF/bcftools merge -m all -f PASS,. --force-samples $DIR/*.vcf.gz |\
$BCF/bcftools plugin fill-AN-AC |\
$BCF/bcftools filter -i 'SUM(AC)>1' > panel_of_normal.vcf
